Tutorial¶
BovineMine is powered by InterMine and provides a user-friendly way to access genomic, proteomic, interaction and literature data. This tutorial is aimed at giving users an introduction to different features of BovineMine.
Overview¶
This section provides a brief overview of the layout for BovineMine.
The navigation panel highlights different functionalities of BovineMine based on what the user is searching for.
- Home - The homepage for BovineMine
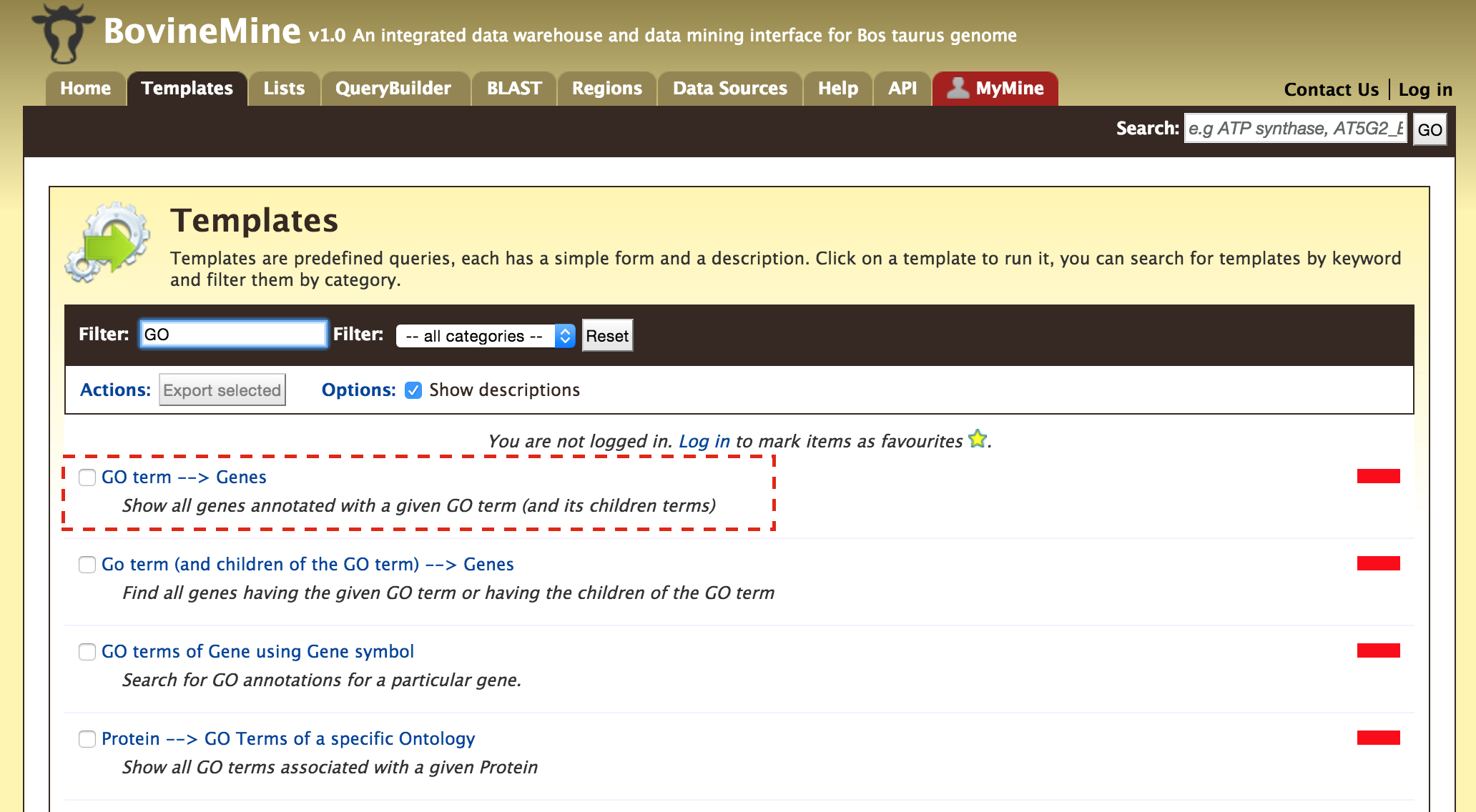
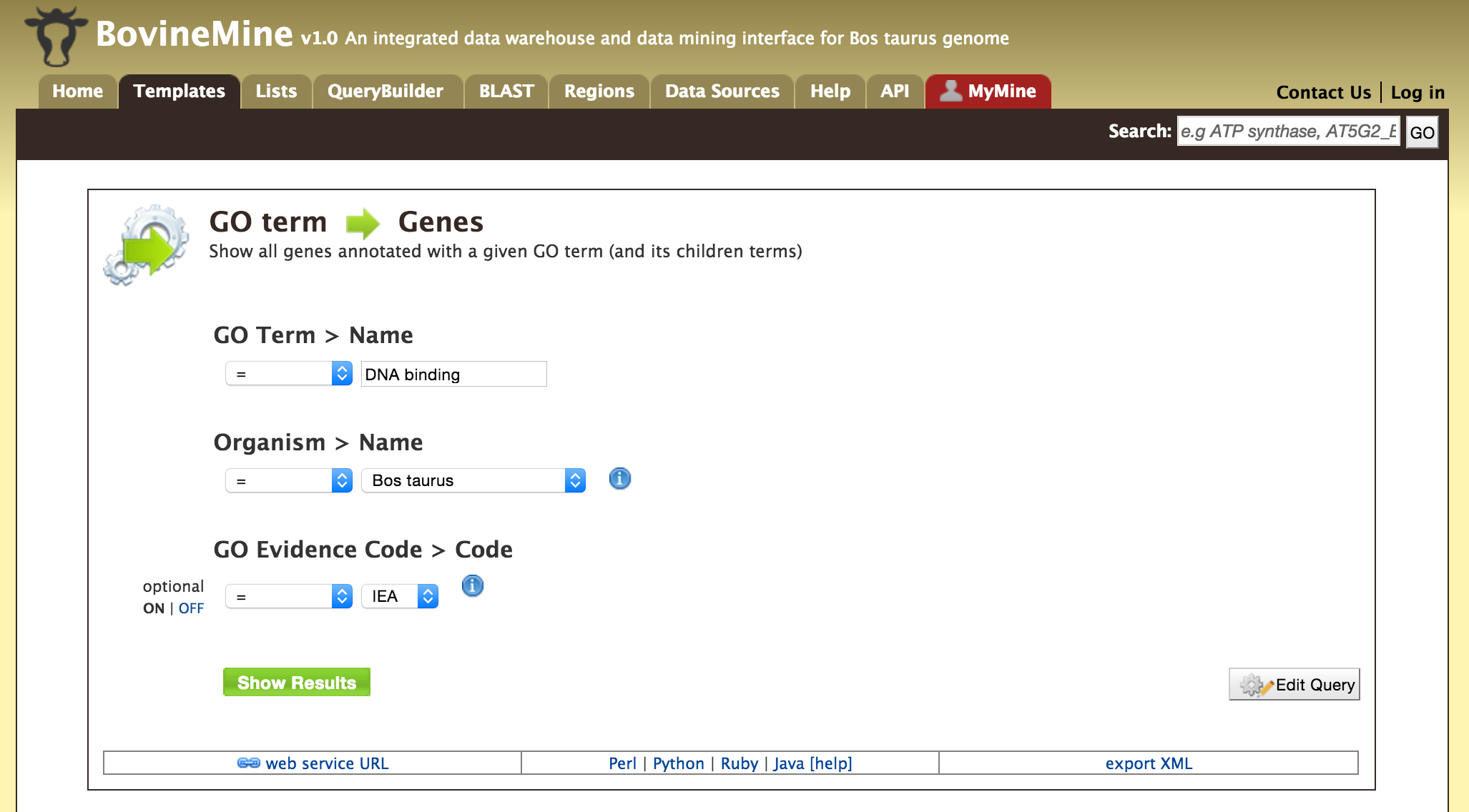
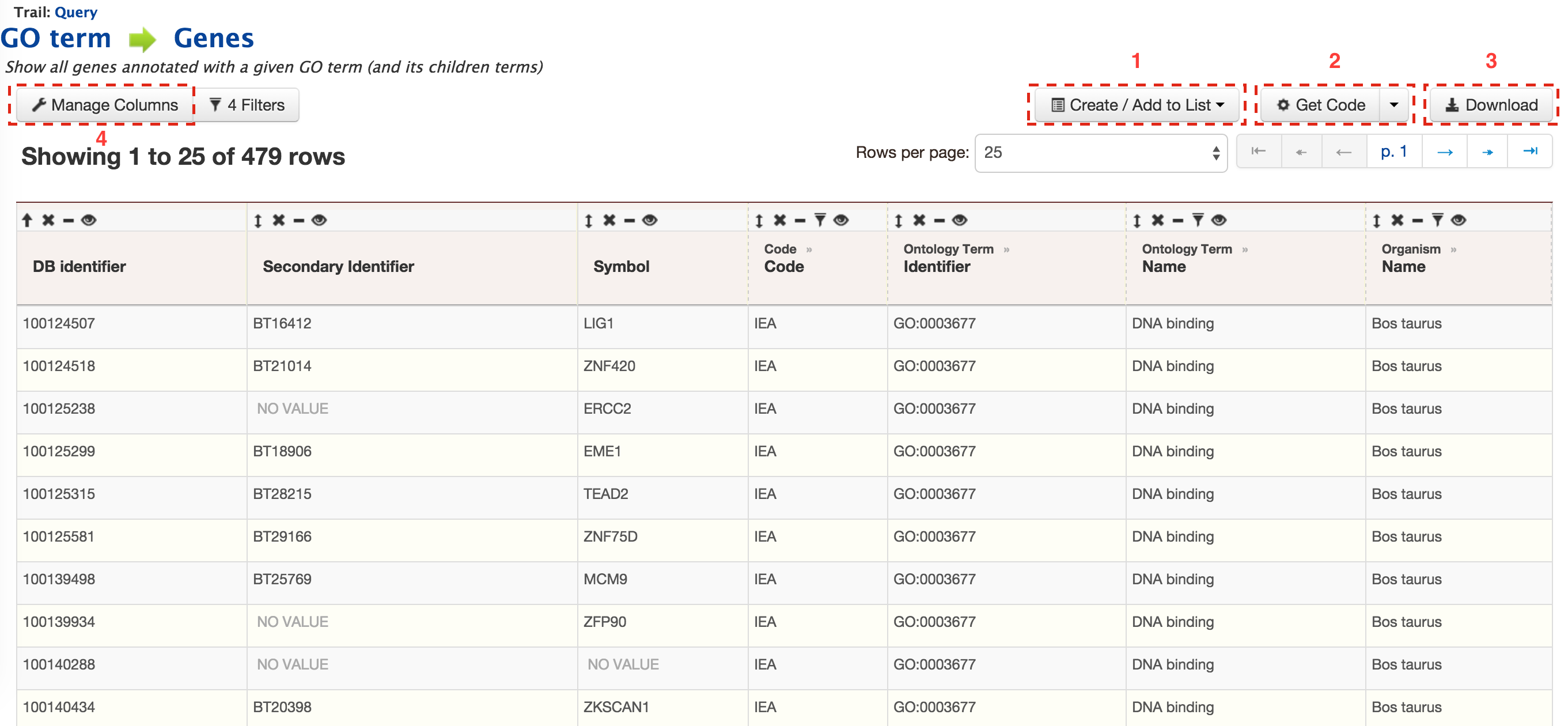
- Templates - Shows the list of templates that users can use based on the nature of their query. Each template is associated with a description which describes what input is expected from the user and what will be the output.
- Lists - The list page offers users a form which they can use to submit or upload a list of genes. Once the list is saved, users can perform enrichment analyses of these genes and export the results.
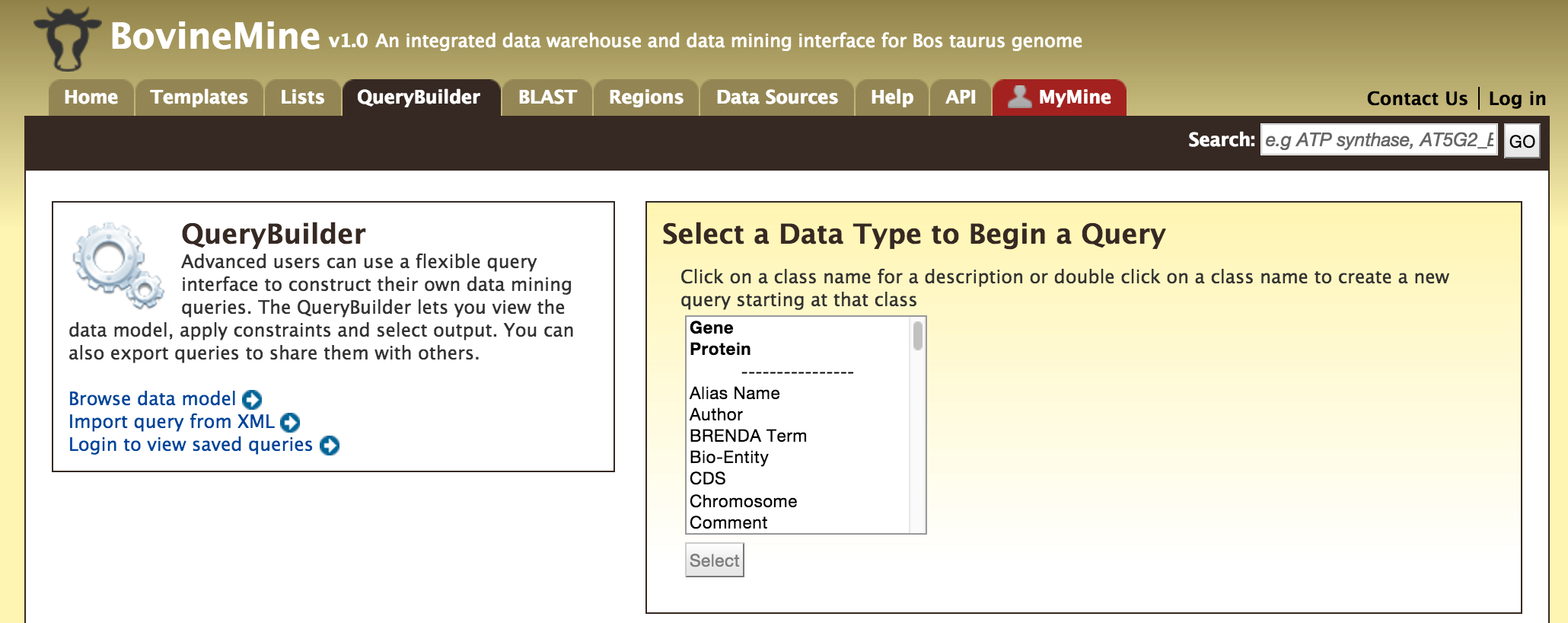
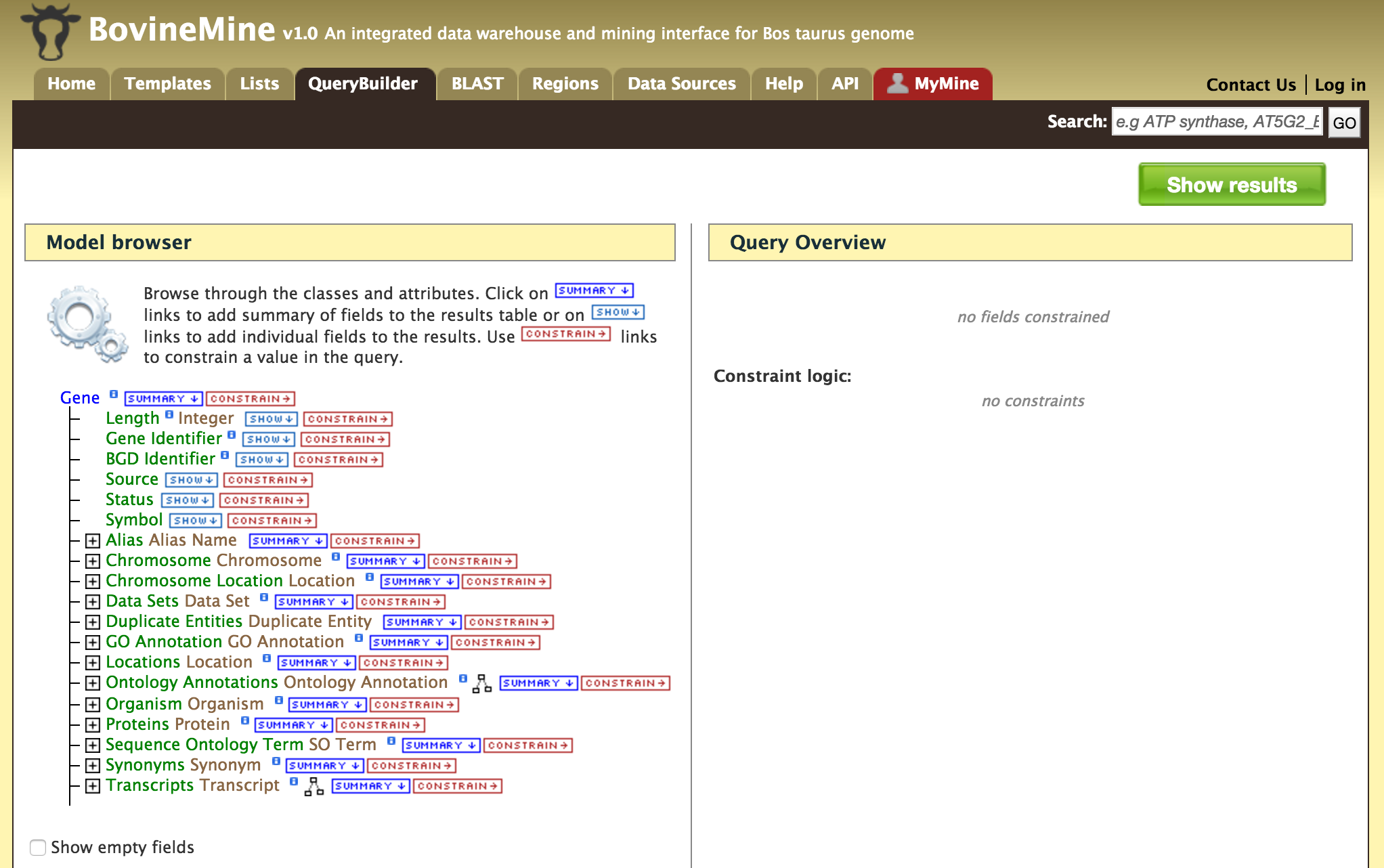
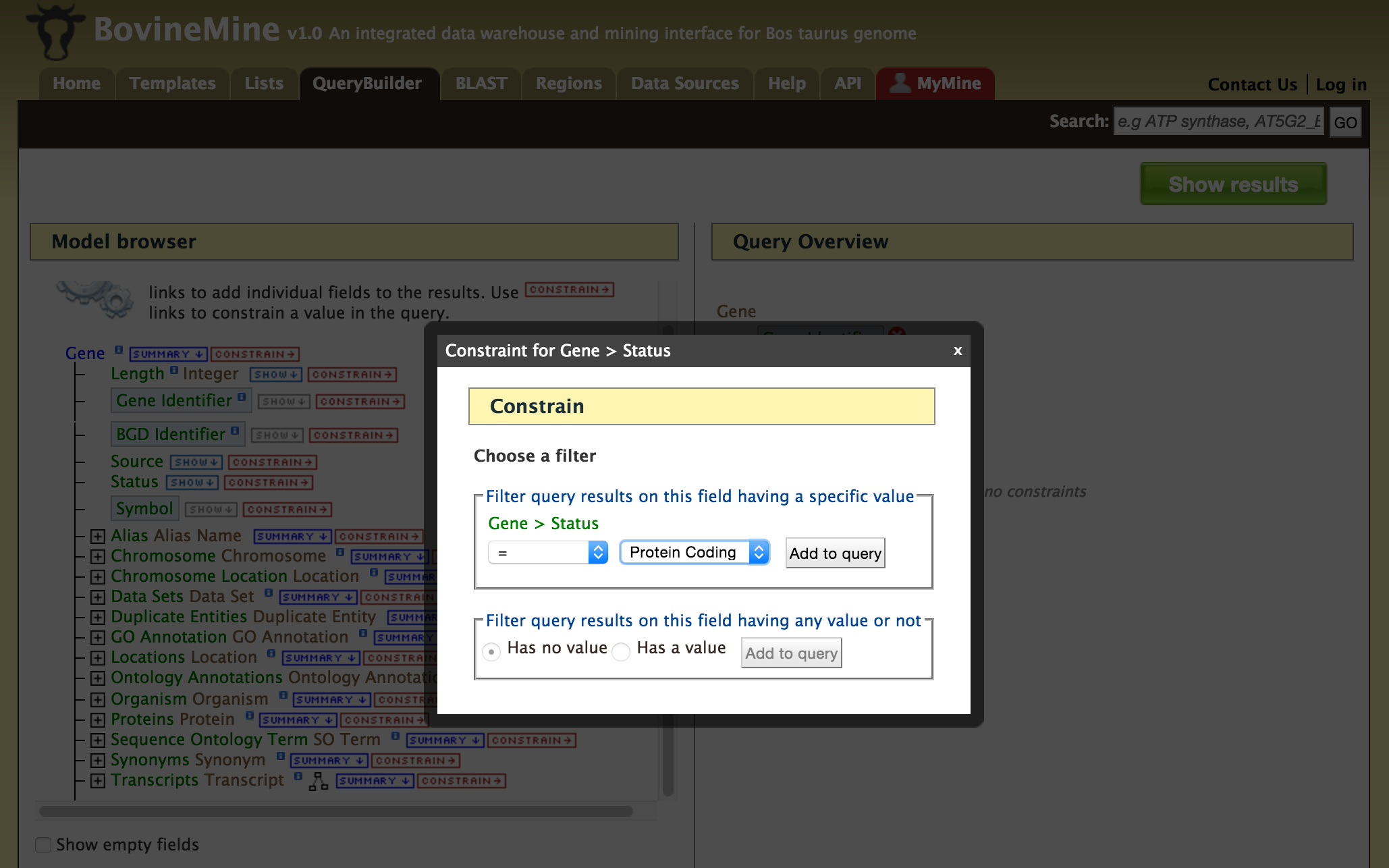
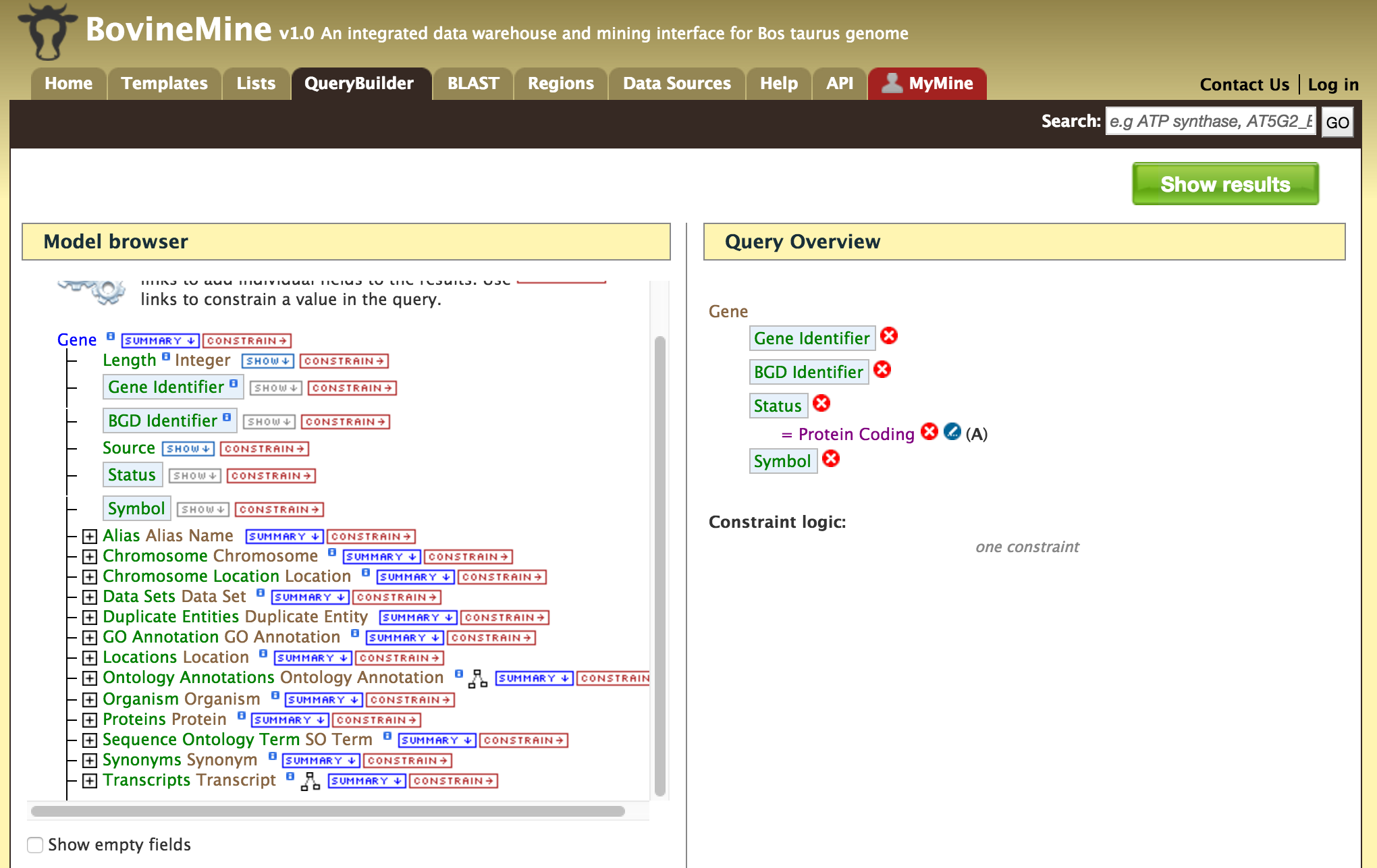
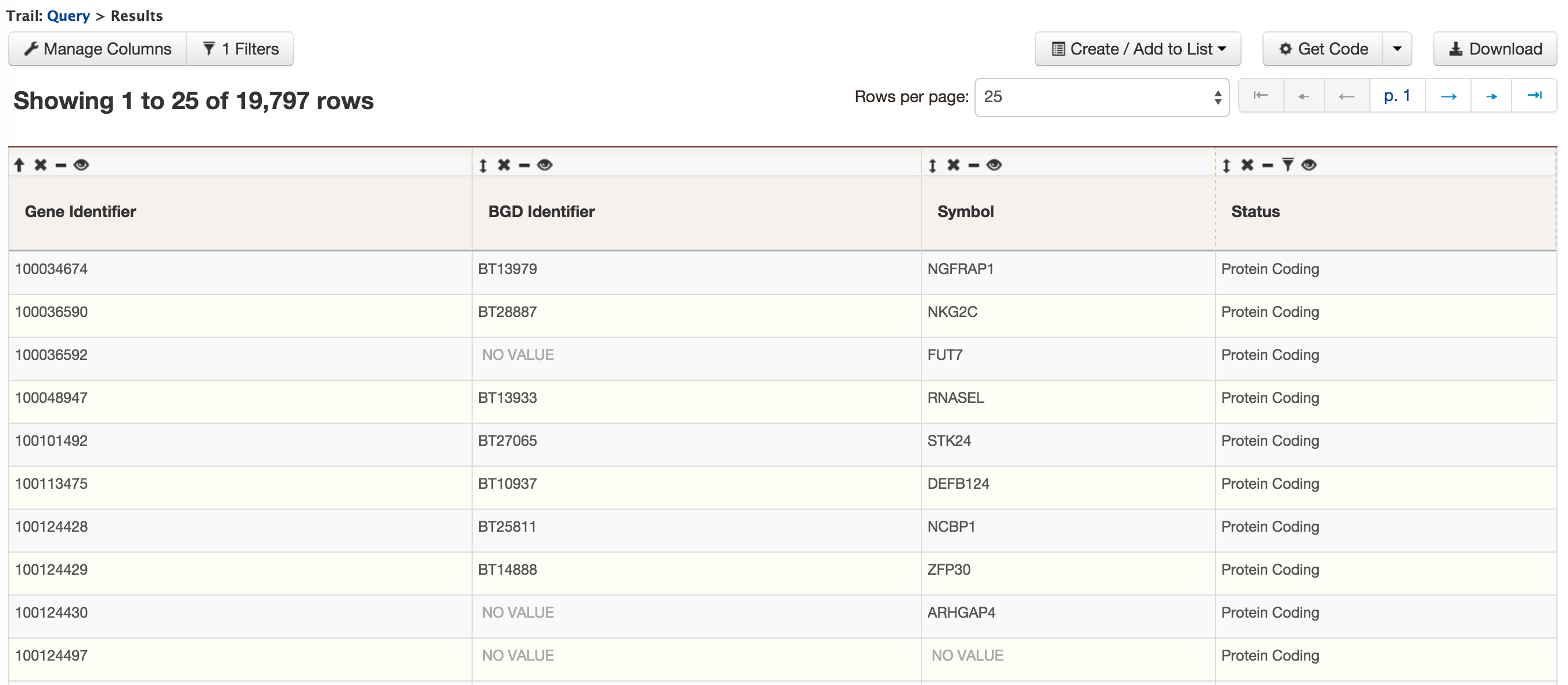
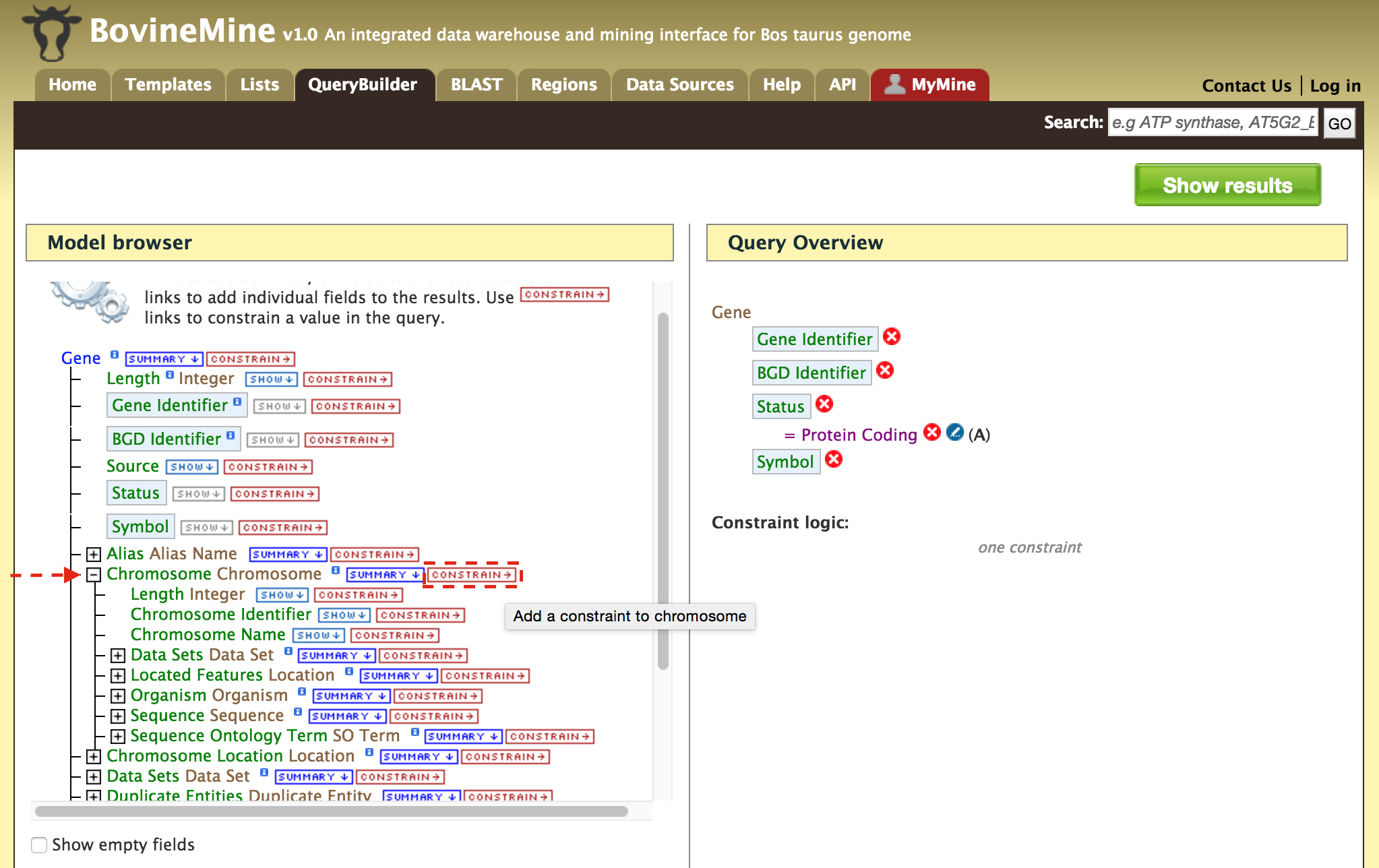
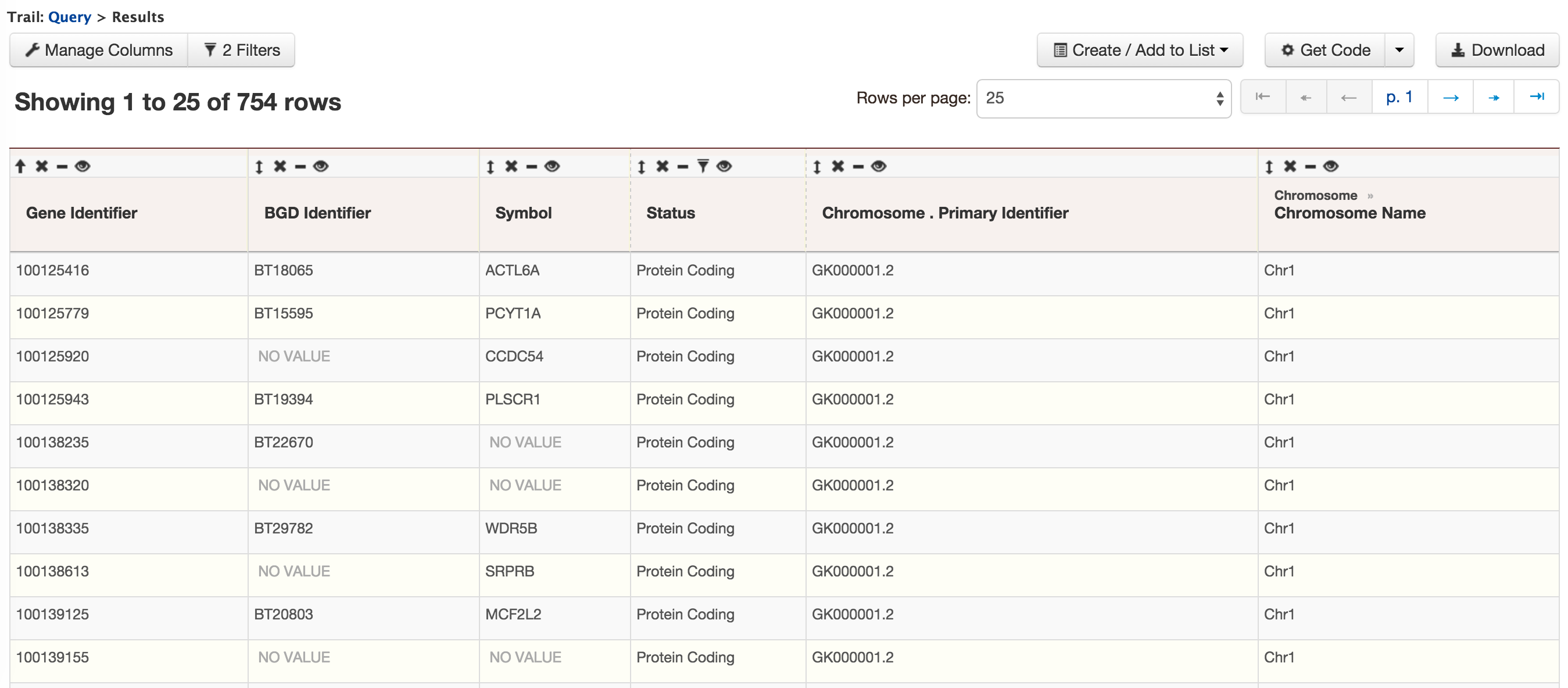
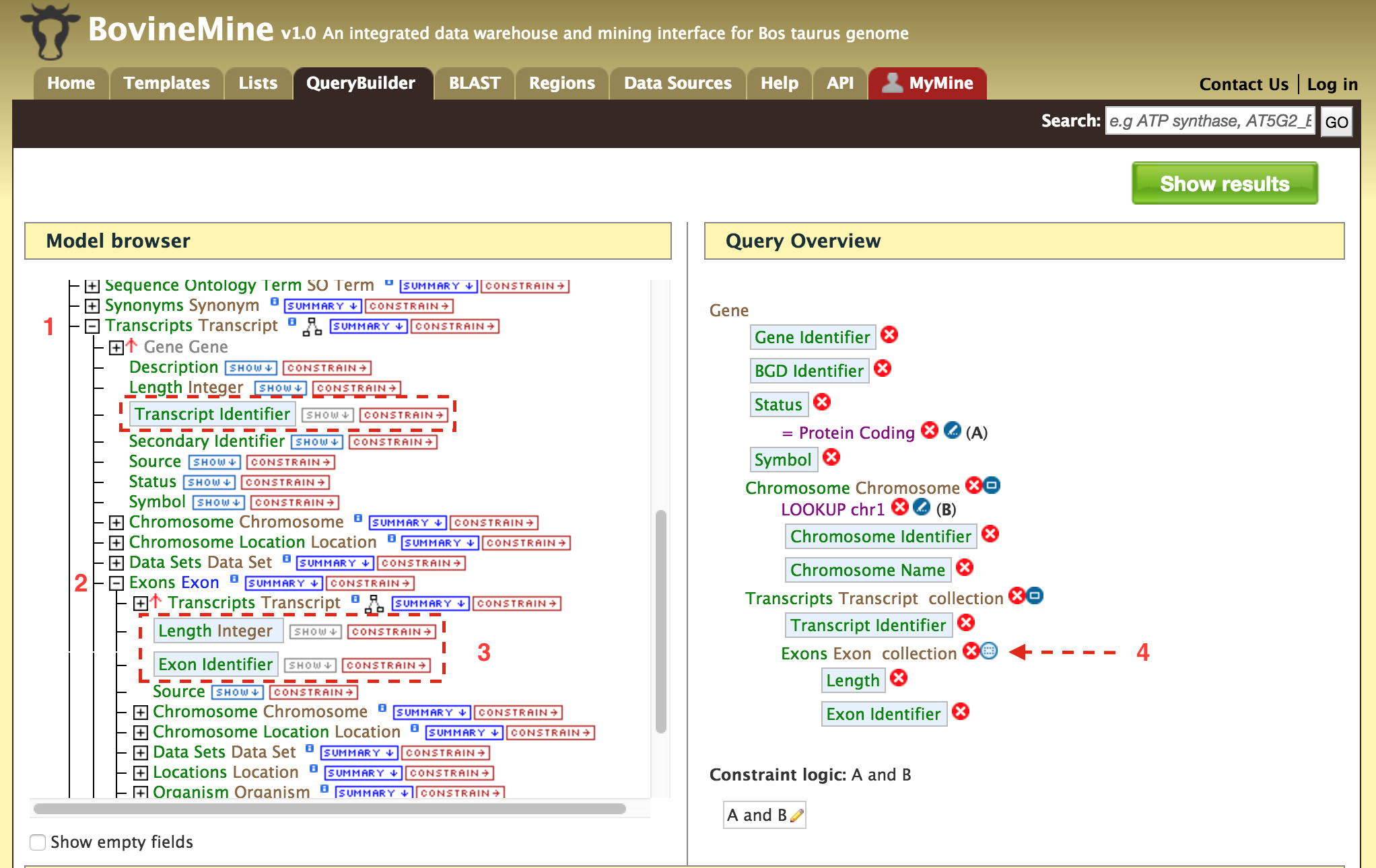
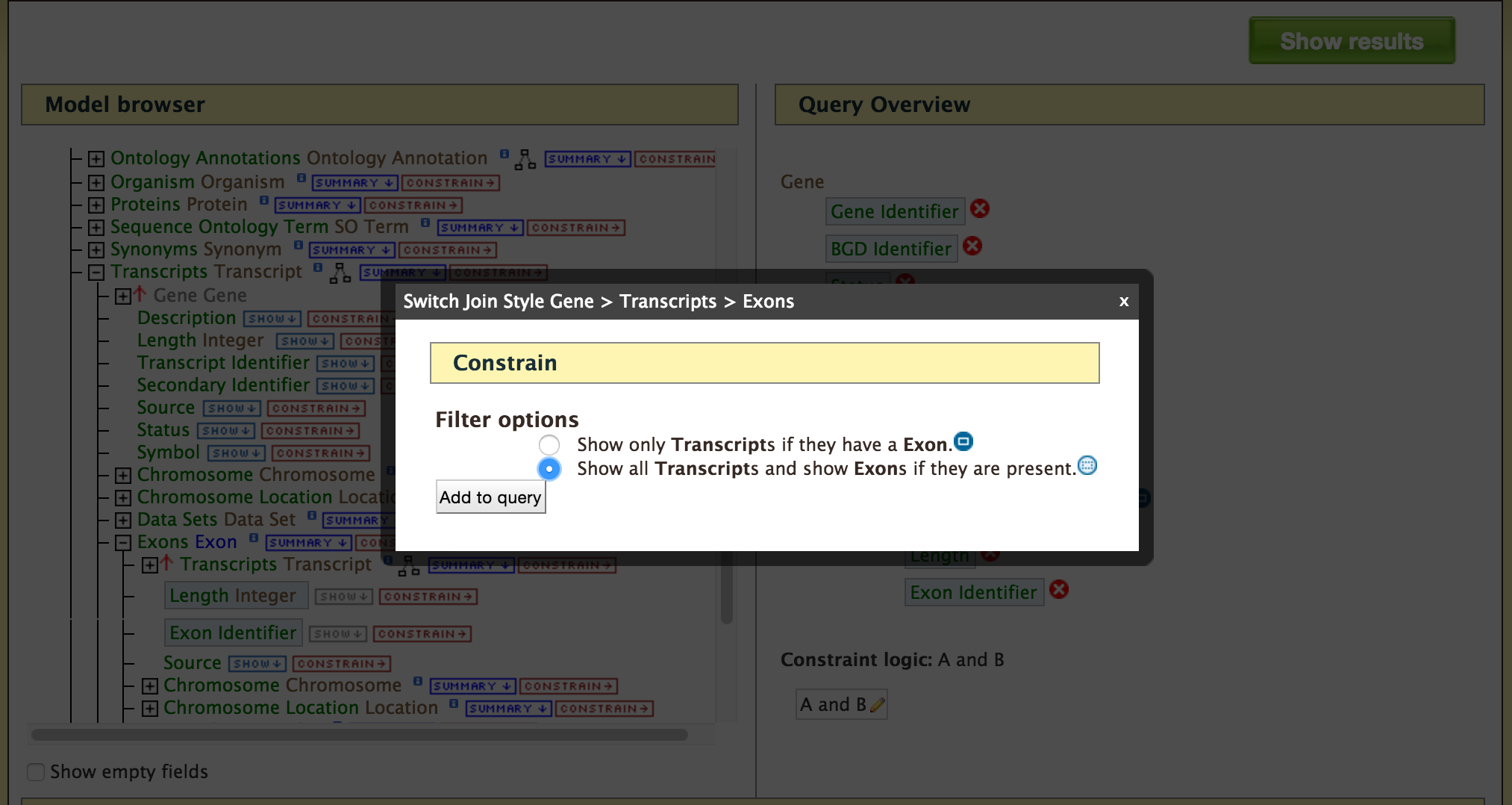
- QueryBuilder - The QueryBuilder is an interface that provides the user to directly interact with the BovineMine data model. Some users might find the QueryBuilder difficult at first, but this tutorial is aimed at familiarizing users on the functionality of QueryBuilder, how to build a custom template and save their own templates.
- BLAST - The BLAST page is where users can search their input sequence(s) against the several databases provided.
- Regions - The Genomic Region Search tool provides a form where users can enter a series of genomic coordinates, specify flanking regions (if any) and fetch all features that fall within the given interval. The result can be exported, saved as a list for further analyses.
- Data Sources - The Data Sources page provides a summary of all the data that is loaded into BovineMine. It lists a combination of datasets from other databases as well as in-house prepared datasets.
- API - The API provides users an option for programmatically accessing BovineMine by using the InterMine API.
- MyMine - The MyMine serves as a portal for users (those who are logged in) for account management. Users can access their saved templates, most recent queries and saved lists.
Report Page¶
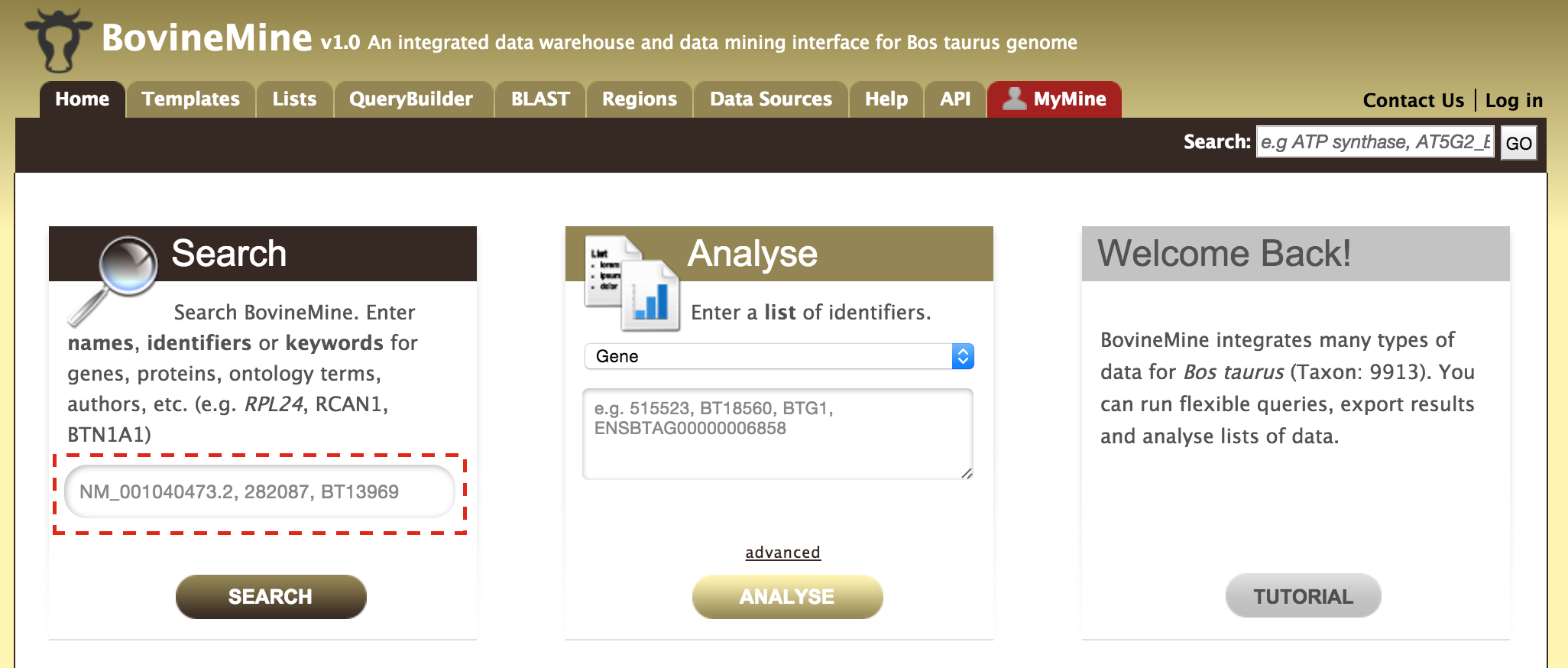
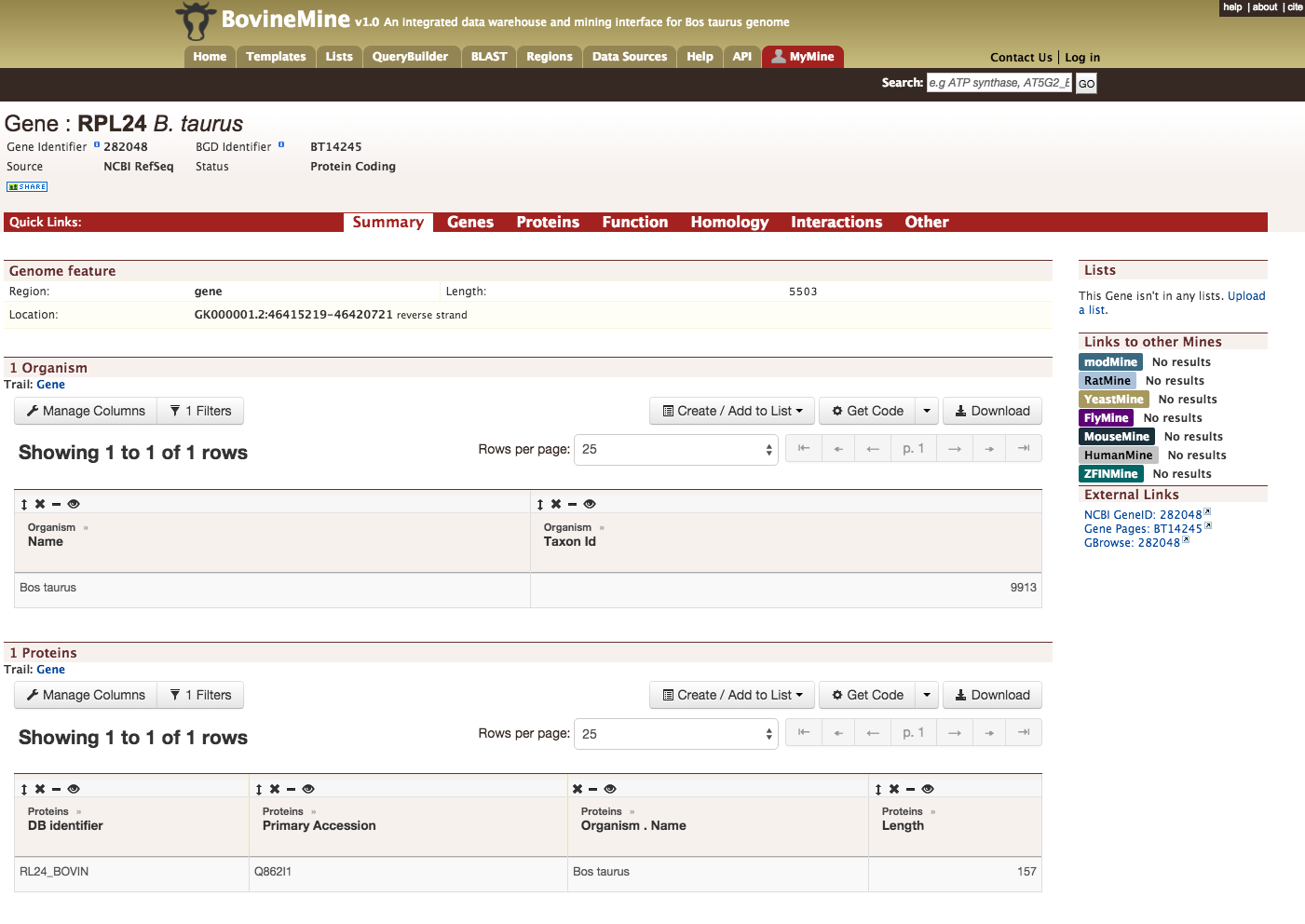
Every query result has a report page and the layout of the report page depends on the data available for a given query. Continuing with the example of rpl24, the report page for this gene is shown below.
The report page provides a complete description for gene rpl24. The header of the report page shows the Gene Identifier (NCBI Gene ID) and status indicating the type of gene, in this case a protein coding gene.
On the right hand side of the report page there are external links that links out to other Mines and databases.
The contents of the report page is divided into categories based on the type of information provided,
Summary
Provides a summary about a gene such as length, chromosome location and strand information. Users can also get the complete FASTA sequence of the gene by clicking on the FASTA tab.
Genes
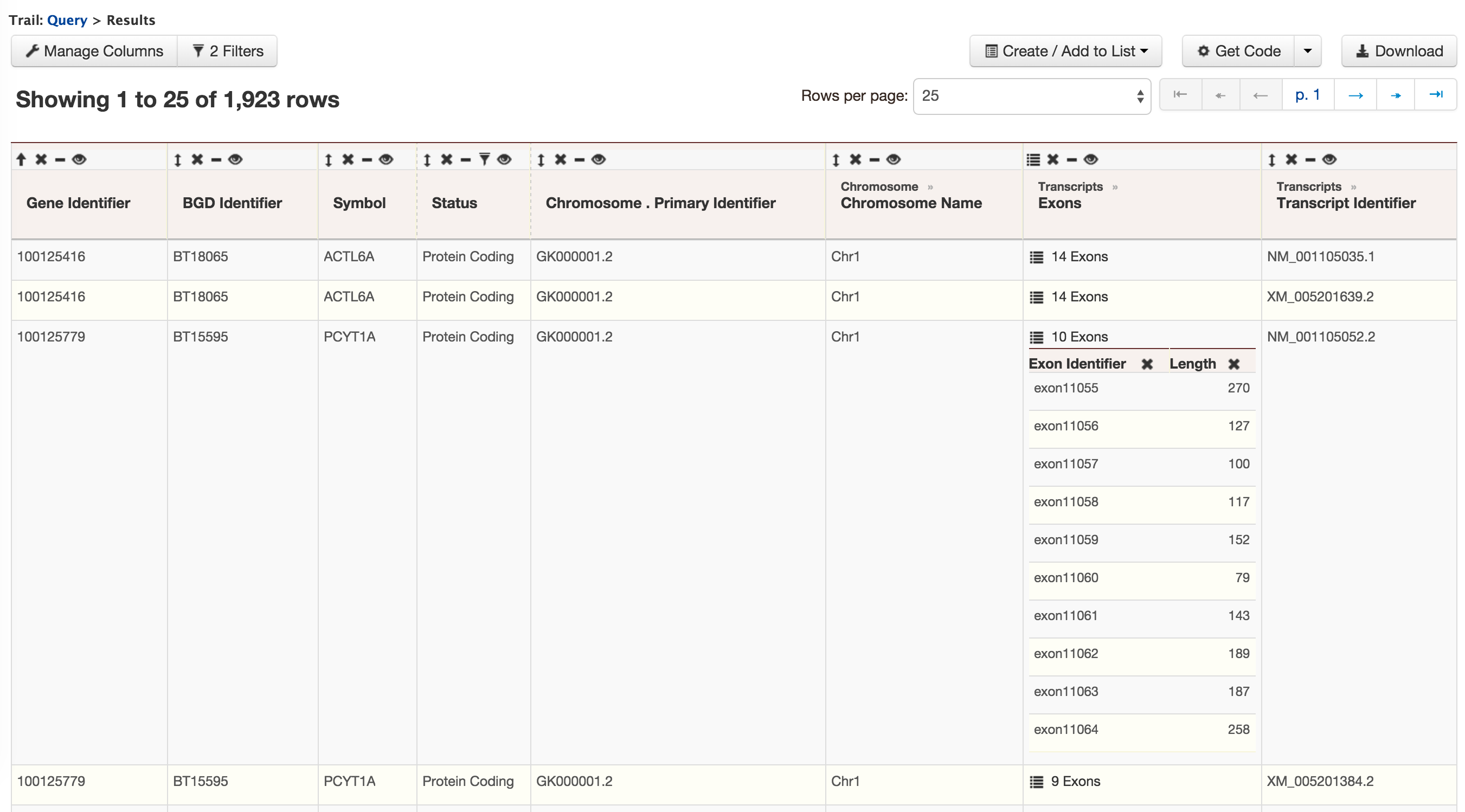
This section provides information about the gene model. It displays all the transcripts and exons for a gene. The FASTA sequence of each transcript or exon can be downloaded by clicking on the FASTA tab. JBrowse is used to visualize the gene model.
Proteins
This section provides information about the protein product of gene rpl24. The comments section gives a brief description about the protein along with the UniProt accession.
Function
This section provides Gene Ontology annotations for a gene and annotations are divided into three categories,
- Biological process
- Molecular function
- Cellular Component
The GO terms are displayed along with the evidence code indicating how the annotations were derived.
Homology
This section provides information on all the homologues for rpl24. The first part shows a summarized view of the homologues in different organisms. The table below gives a detailed information about the homologue, the type of homologue and from which dataset the information was obtained.
Interaction
This section provides information about interactions. For rpl24 there are no interaction information available but for genes that do have interaction information, a network is displayed showing all interactors for the current gene. The network displayer is a Cytoscape plugin.
Others
This section provides additional information such as,
- Child features – lists all the features that are sub features of the current gene
- Flanking regions – lists all the features flanking the current gene
- Overlapping features – lists all the features that overlap with current gene
- Pathways – lists all the pathways in which the current gene is a part of
- Publications – Publications related to the current gene
BLAST¶
Users can perform BLAST against the Bos taurus genomic, CDS or protein sequences using the BLAST page.
Genomic Region Search¶
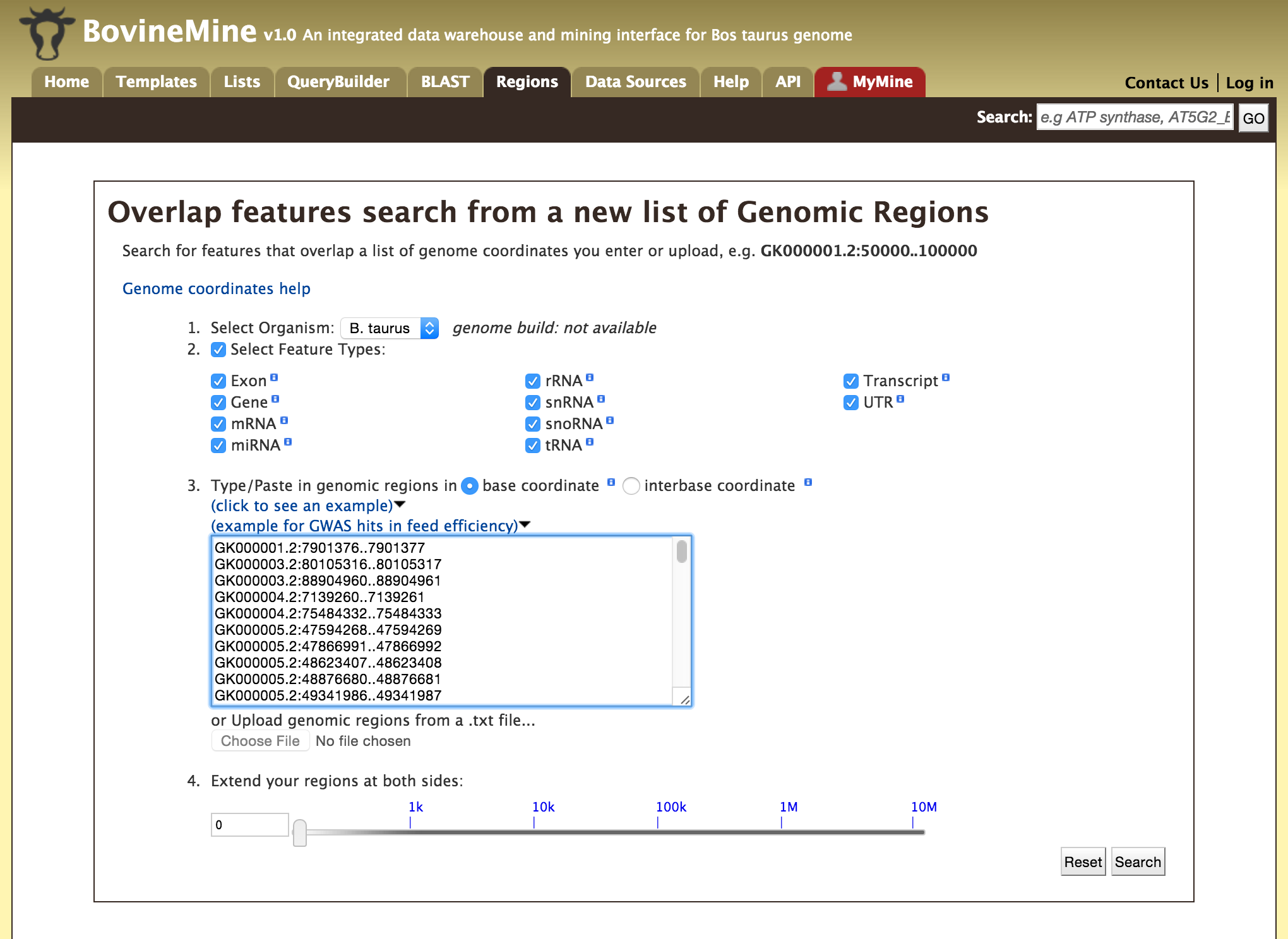
The Genomic Region Search is a tool to fetch features that are within a given set of genomic coordinates or to fetch features that are within a given number of bases flanking a given set of genomic coordinates.
The coordinates have to be of the format,
chromosome_name:start..end
chromosome_name:start-end
chromosome_name start end
Click on click to see an example for a representative set of genomic coordinates.
Users can extend the regions on either side of a given coordinate using the slider or using the textbox.
Users can also select the type of coordinate system they would like to use: base coordinate or interbase coordinate.
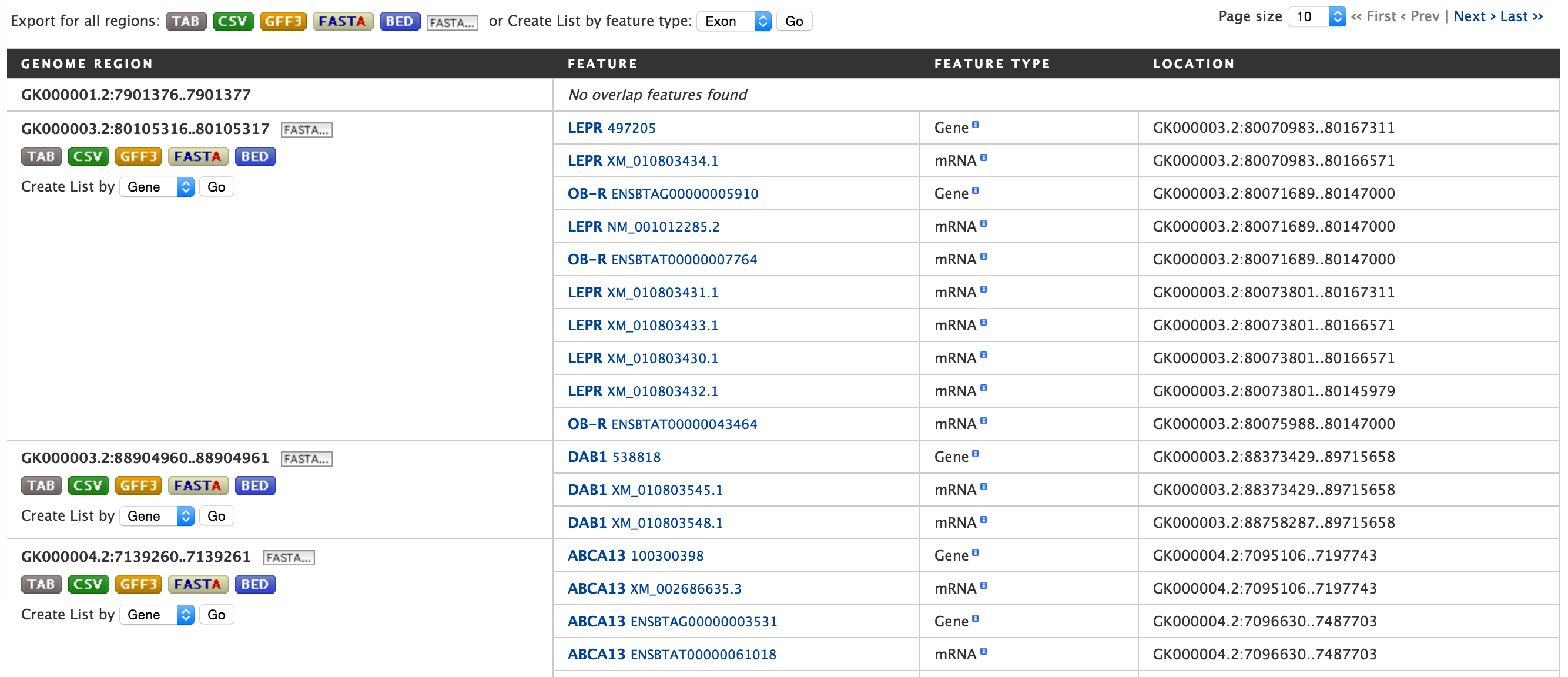
Lets try and example. Click on the click to see an example and extend the region search by 500 bp and click on Search. The result page will give a list of features that are present in each of the genomic interval provided in the input.
The results can be exported as tab-delimited and comma-separated values. If the results have genomic features then they can be exported in GFF3 or BED format. Users can also export FASTA sequences of the features. If users are interested in creating a list of particular features from the result page then they can filter based on feature type, shown in red box, and click on ‘Go’.
Lists¶
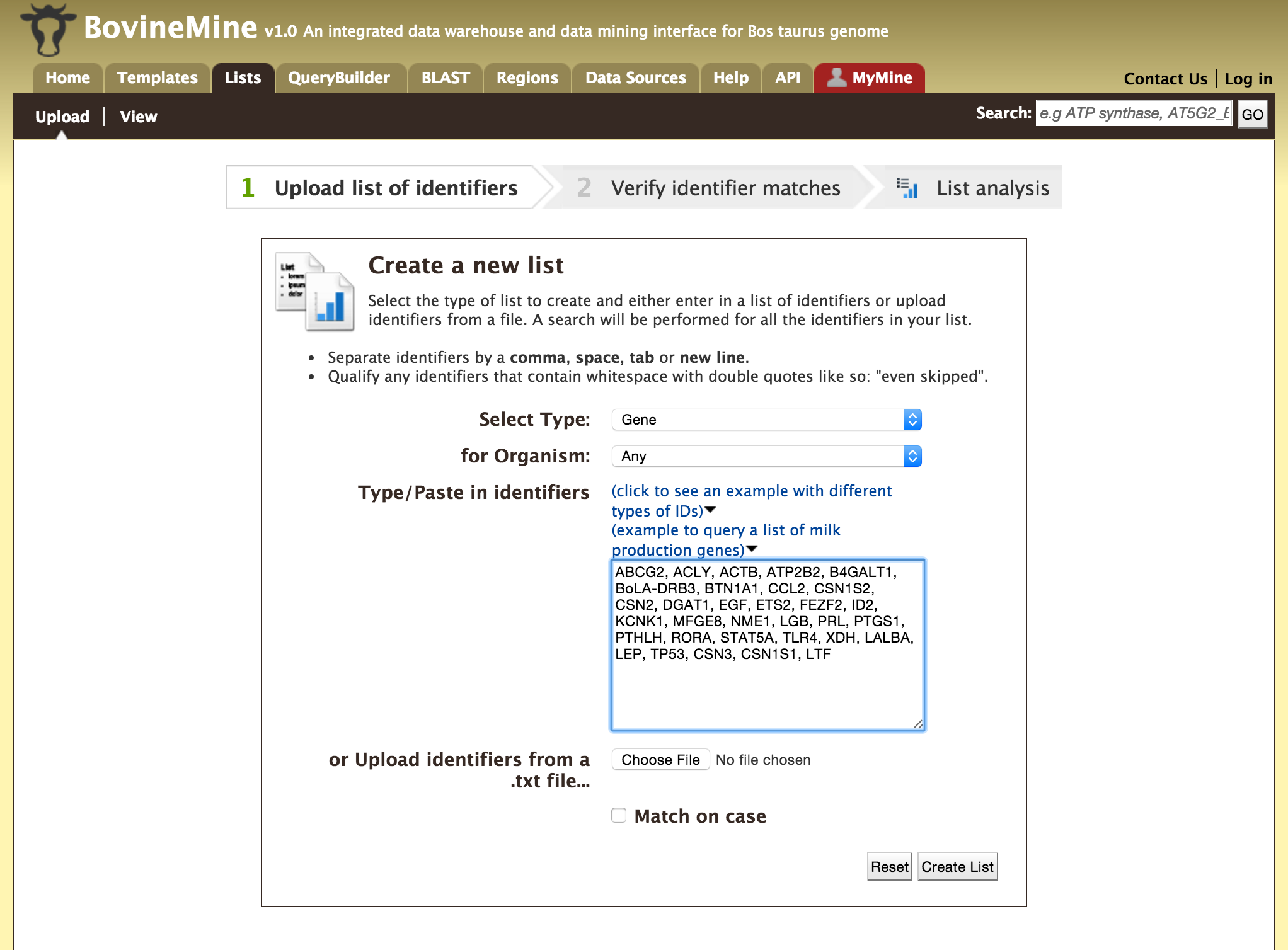
Users can create a list of features. The input can either be gene IDs, transcript IDs, gene symbols, etc.
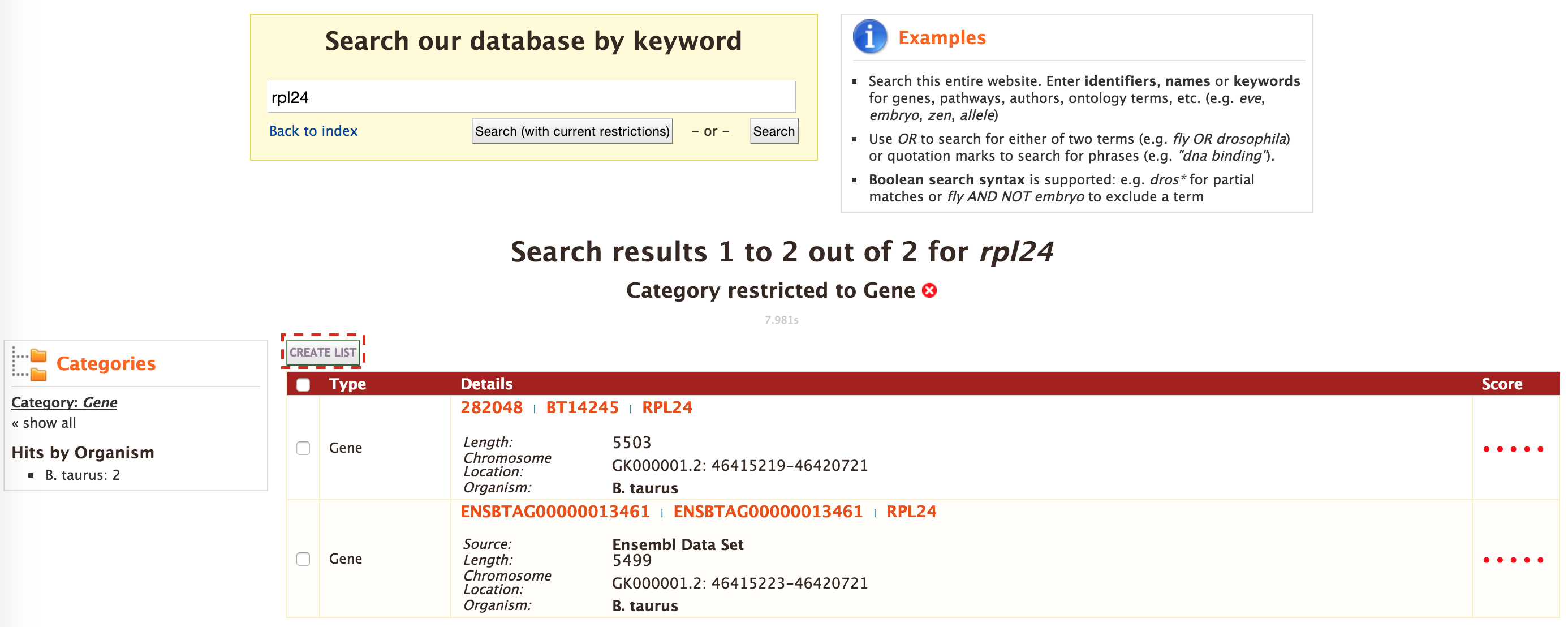
The list tool tries to lookup the query throughout the database and will attempt to convert the identifiers to the type selected in the list ‘Select Type’ option. Lets try the examples provided. Click on ‘Click to see example’ link (highlighted in red box) and click on ‘Create List’.
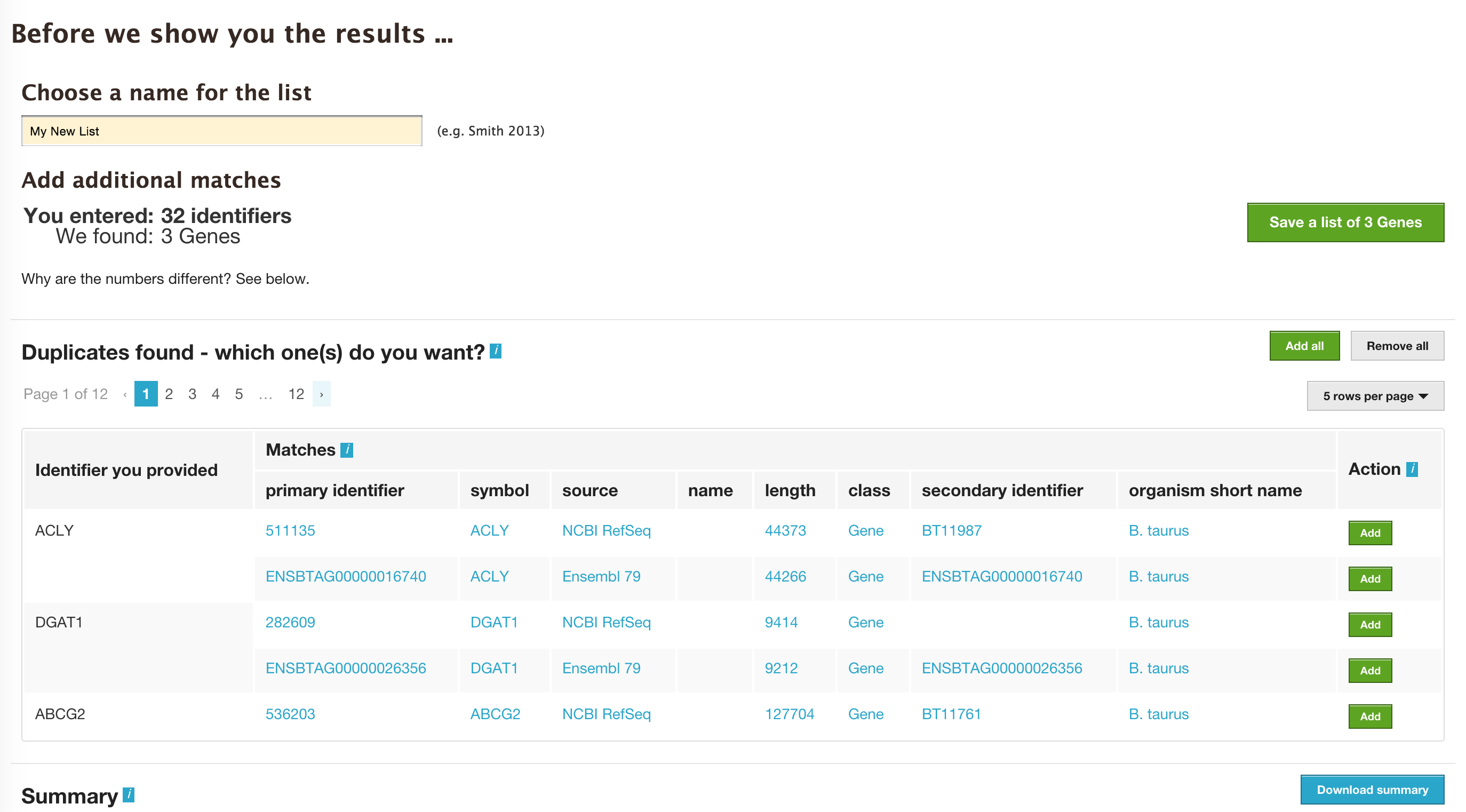
The list tool does a lookup of the identifiers and shows you the results. If there are any duplicates, users can decide to add the relevant entries individually.
The summary section provides information regarding those identifiers that had a direct hit without any duplicates.
Click on ‘Add all’ and then click on ‘Save a list of 62 genes’.
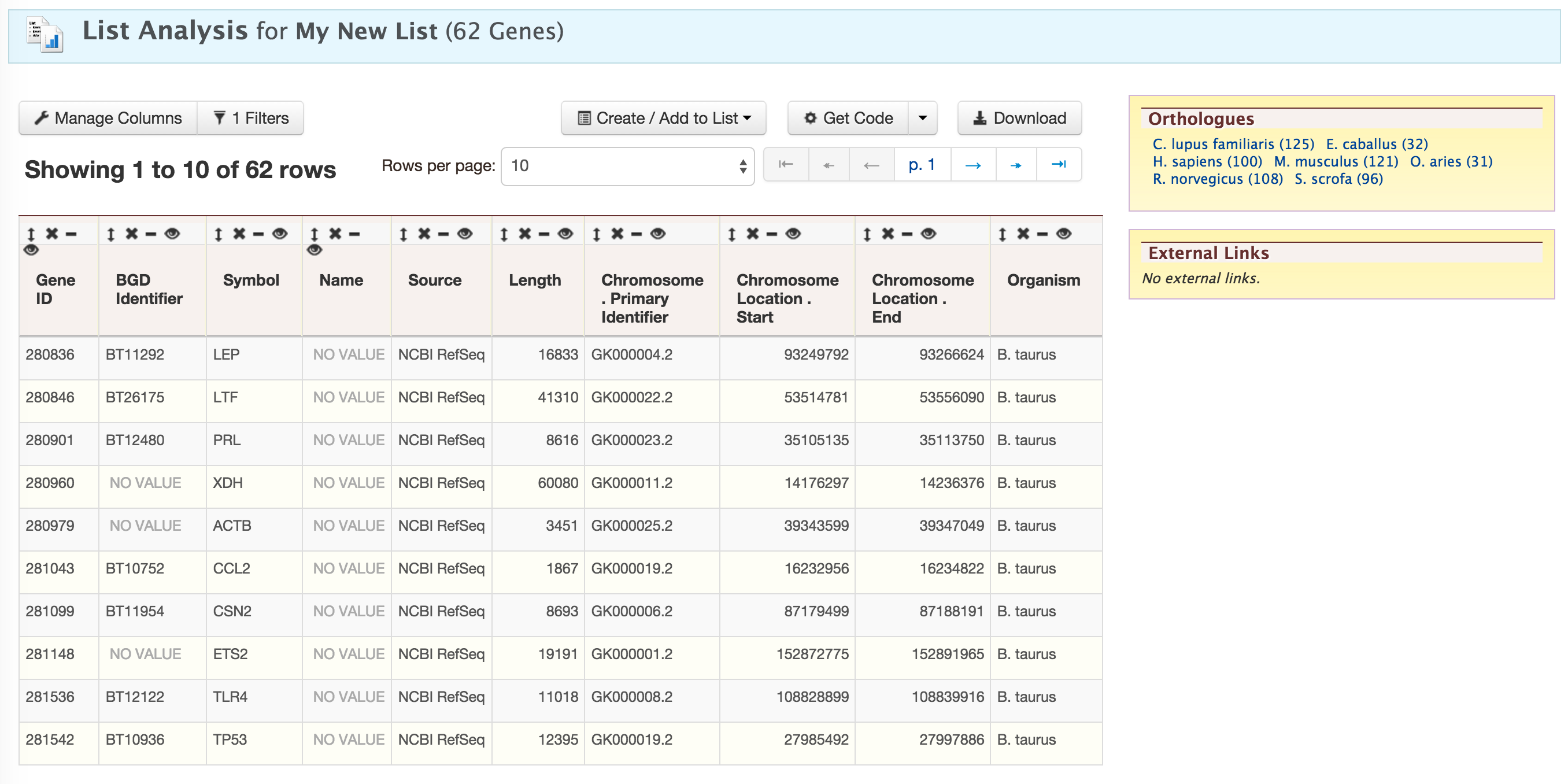
This will take users to a List Analysis page,
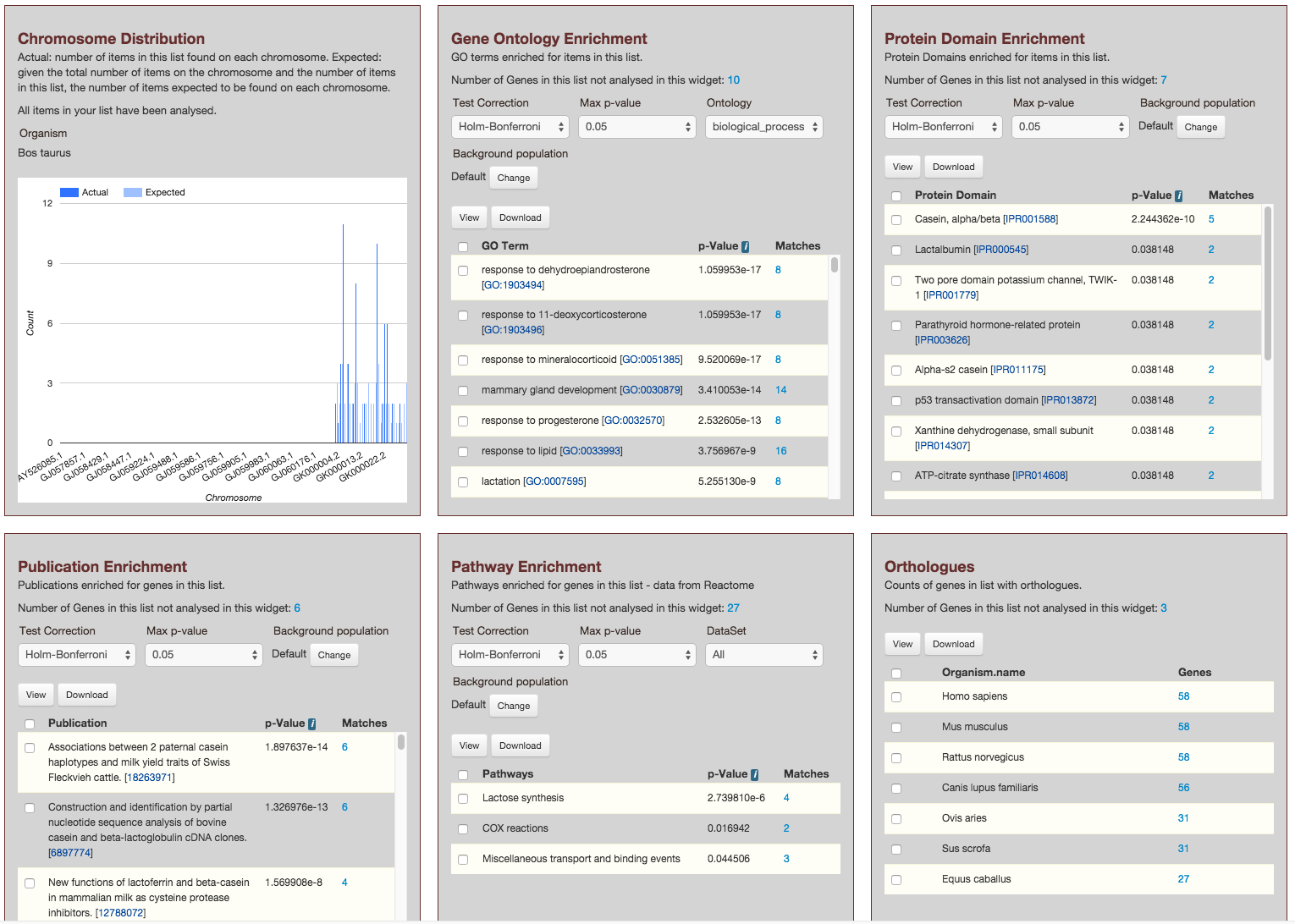
This page provides users with widgets to perform analyses on gene lists that they have created.
Currently available widgets are,
- Chromosome Distribution
- Gene Ontology Enrichment
- Protein Domain Enrichment
- Publication Enrichment
- Pathway Enrichment
- Orthologues
MyMine¶
Users can create an account on BovineMine which enables them to store their lists, custom templates and keep track of their sessions. To create a login, click on the ‘Log in’ on the top-right corner of any page on BovineMine.
New users can create a new account with the username being their Email ID.
BovineMine provides MyMine for users to manage their lists, queries, templates, and account details.
- Lists – lists saved by the user.
- History of queries by user – shows a list of most recent queries performed by the user.
- Templates – Templates created by the user or existing templates that are marked as ‘favorite’ by the user.
- Password change – change the password for the user’s account.
- Account details – for updating user preferences.
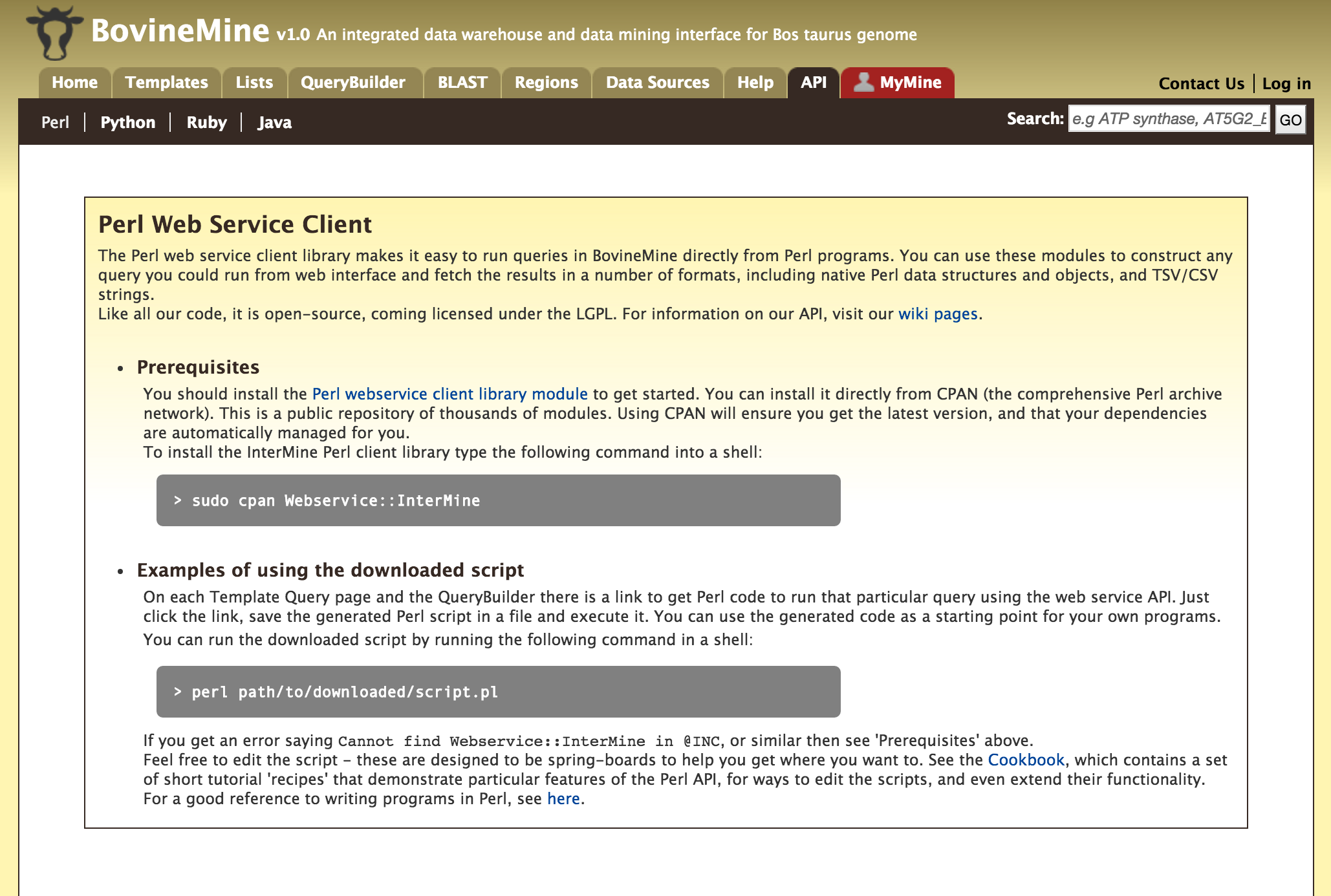
API¶
For users who would like to programmatically access BovineMine they can make use of the InterMine API.
Perl, Python, Ruby and Java are the 4 languages in which the InterMine API is available. For more information on the details on the API visit the InterMine Wiki.
Data Sources¶
Provides a description of the datasets that are integrated into BovineMine along with the location from which these datasets were downloaded, their version or release, citations wherever applicable and any additional comments.